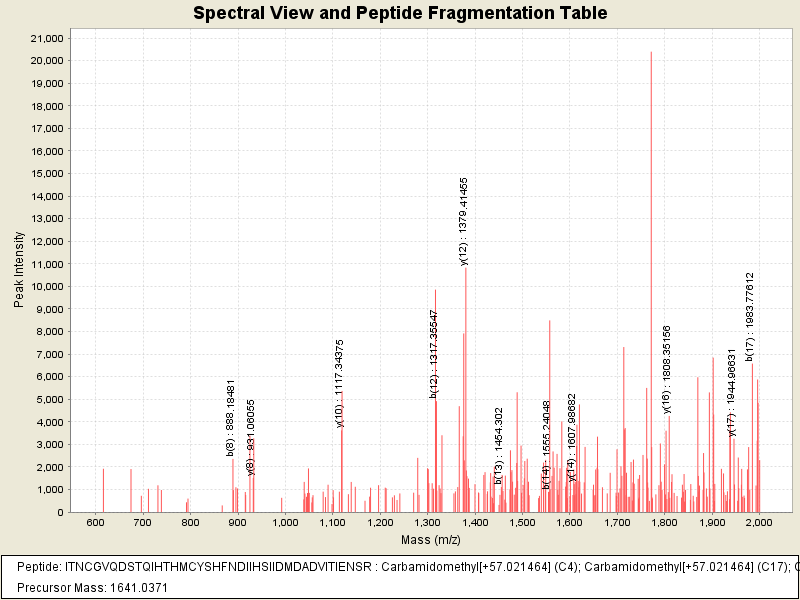

Observed mass of peptide: 1641.037109375, (+3)
Observed intensity of peptide: 3557137.0
Sequest Xcorr: 2.7984042167663574
| # | b++ | b | Residue | y | y++ | # |
|---|---|---|---|---|---|---|
| 1 | 57.5499 | 114.0919 | I | 4917.2345 | 2459.1212 | 42 |
| 2 | 108.0737 | 215.1396 | T | 4804.1505 | 2402.5792 | 41 |
| 3 | 165.0952 | 329.1825 | N | 4703.1028 | 2352.0553 | 40 |
| 4 | 245.1105 | 489.2132 | C +57.0215 | 4589.0599 | 2295.0338 | 39 |
| 5 | 273.6212 | 546.2346 | G | 4429.0292 | 2215.0185 | 38 |
| 6 | 323.1554 | 645.303 | V | 4372.0077 | 2186.5078 | 37 |
| 7 | 387.1847 | 773.3616 | Q | 4272.9393 | 2136.9736 | 36 |
| 8 | 444.6982 | 888.3886 | D | 4144.8807 | 2072.9443 | 35 |
| 9 | 488.2142 | 975.4206 | S | 4029.8538 | 2015.4308 | 34 |
| 10 | 538.7381 | 1076.4683 | T | 3942.8218 | 1971.9148 | 33 |
| 11 | 602.7673 | 1204.5269 | Q | 3841.7741 | 1921.391 | 32 |
| 12 | 659.3094 | 1317.6109 | I | 3713.7155 | 1857.3617 | 31 |
| 13 | 727.8388 | 1454.6698 | H | 3600.6314 | 1800.8196 | 30 |
| 14 | 778.3627 | 1555.7175 | T | 3463.5725 | 1732.2902 | 29 |
| 15 | 846.8921 | 1692.7764 | H | 3362.5248 | 1681.7663 | 28 |
| 16 | 912.4124 | 1823.8169 | M | 3225.4659 | 1613.2369 | 27 |
| 17 | 992.4277 | 1983.8476 | C +57.0215 | 3094.4254 | 1547.7166 | 26 |
| 18 | 1073.9594 | 2146.9109 | Y | 2934.3948 | 1467.7013 | 25 |
| 19 | 1117.4754 | 2233.9429 | S | 2771.3315 | 1386.1696 | 24 |
| 20 | 1186.0048 | 2371.0019 | H | 2684.2994 | 1342.6536 | 23 |
| 21 | 1259.539 | 2518.0703 | F | 2547.2405 | 1274.1242 | 22 |
| 22 | 1316.5605 | 2632.1132 | N | 2400.1721 | 1200.59 | 21 |
| 23 | 1374.074 | 2747.1402 | D | 2286.1292 | 1143.5685 | 20 |
| 24 | 1430.616 | 2860.2242 | I | 2171.1022 | 1086.055 | 19 |
| 25 | 1487.1581 | 2973.3083 | I | 2058.0182 | 1029.513 | 18 |
| 26 | 1555.6875 | 3110.3672 | H | 1944.9341 | 972.971 | 17 |
| 27 | 1599.2035 | 3197.3992 | S | 1807.8752 | 904.4415 | 16 |
| 28 | 1655.7456 | 3310.4833 | I | 1720.8431 | 860.9255 | 15 |
| 29 | 1712.2876 | 3423.5674 | I | 1607.7591 | 804.3834 | 14 |
| 30 | 1769.8011 | 3538.5943 | D | 1494.675 | 747.8414 | 13 |
| 31 | 1843.3188 | 3685.6297 | M +15.9949 | 1379.648 | 690.3279 | 12 |
| 32 | 1900.8323 | 3800.6567 | D | 1232.6126 | 616.8102 | 11 |
| 33 | 1936.3508 | 3871.6938 | A | 1117.5857 | 559.2968 | 10 |
| 34 | 1993.8643 | 3986.7207 | D | 1046.5486 | 523.7782 | 9 |
| 35 | 2043.3985 | 4085.7892 | V | 931.5216 | 466.2647 | 8 |
| 36 | 2099.9405 | 4198.8732 | I | 832.4532 | 416.7305 | 7 |
| 37 | 2150.4644 | 4299.9209 | T | 719.3691 | 360.1885 | 6 |
| 38 | 2207.0064 | 4413.005 | I | 618.3215 | 309.6646 | 5 |
| 39 | 2271.5277 | 4542.0476 | E | 505.2374 | 253.1226 | 4 |
| 40 | 2328.5492 | 4656.0905 | N | 376.1948 | 188.6013 | 3 |
| 41 | 2372.0652 | 4743.1225 | S | 262.1519 | 131.5798 | 2 |
| 42 | 2450.1157 | 4899.2237 | R | 175.1198 | 88.0638 | 1 |